Chromatin represents the combination of both the DNA and proteins in a cell nucleus. This combined material can assume specific configurations. This chromatin architecture is currently viewed as having a critical role in controlling gene expression and DNA replication. The primary protein elements of chromatin, or histones, compact DNA and assist in mitosis. Epigenetic changes occur by complex interactions between environmental influences and the underlying protein structure and DNA of a genome, which can alter the configuration of chromatin or change how segments interact in their three-dimensional environment. Examples include DNA methylation and alternations to chromatin architecture through histone modification. It is believed that most epigenetic modifications to underlying DNA sequences, either through promotion of genetic expression or silencing of…Read More
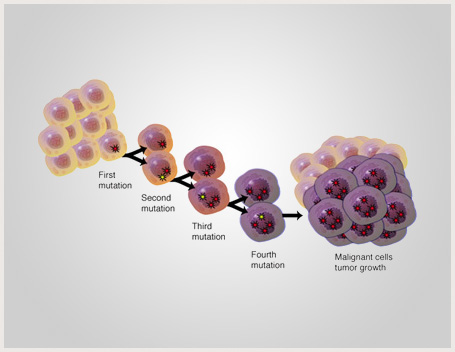
New research is demonstrating that the etiology of cancer is complex. Epigenetic modifications of the genomes of cancer cells appear to be quite important. These are changes that do not necessarily reflect the exact DNA sequences within a cell, but alter, by various means, the expression of the underlying code. It is increasingly apparent that epigenetic modification may be as important as genetic mutation in the transformation of a normal cell to a cancer cell. This does not change the fact that many cancers are induced by infectious disease which itself can be considered an epigenetic variable. Such epigenetic mechanisms are being successfully defined. These include DNA methylation, arguably the most researched marker, by which the methylation profile can be linked to tumorigenesis as…Read More
New scientific findings involving epigenetics are having a profound impact on our understanding of evolutionary mechanisms. It is becoming increasingly apparent that epigenetic modifications play a critical role in the regulation of gene expression. In so doing, they produce phenotypic changes. It is becoming abundantly clear that these influences are an important factor in our own human experiences, influencing life span, mood disorders, immunological status, obesity or tendencies towards the variety of chronic illnesses that have afflicted our human experience. Although it has until recently been assumed that these epigenetic marks are cleared in each generation and then re-established by reiteration of similar environmental influences, it is now manifest that this is not always the case. The term ‘transgenerational epigenetic inheritance’…Read More

This is the first of a five part series discussing epigenetics. In science, what is old can once again become the new. To better understand the definition of epigenetics, we must look back at the time before Darwin. Charles Darwin proposed his version of natural selection and heritable descent by gradual modification in 1859, but before that, many theories had been considered before creationism. One leading evolutionary proponent was Jean-Baptiste Chevalier do Lamarck. He was the protégé of the Comte de Buffon who had believed that living organisms can vary over time and their experiences in the environment may affect them and their progeny. Lamarck proposed the concept of the inheritance of acquired characteristics as just this sort of mechanism. He believed that…Read More
Metagenomic evaluation of any environment, ecology or tissue sample requires extensive computational power in order to properly assess large volumes of sequence data. This is required to mobilize an accurate representation of genetic diversity in a sample. New techniques in de novo metagenomic assembly effectively filter the total amount of data to be analyzed; yet, vast amounts of computer power are still required. New approaches to that analysis and changes in the filtering methods have enabled improved methods of metagenome assembly of a more accurate characterization of the full range of microbial life in a sample. Complex software is necessary since in conventional genome analysis, only one species is being analyzed. However, metagenomic sample analysis, the required metagenome assemblers use algorithms to additionally…Read More

